Development/Vampir and VampirServer: Difference between revisions
No edit summary |
|||
| Line 80: | Line 80: | ||
bwunicluster$ vampir # Starting the Qt Application Vampir GUI |
bwunicluster$ vampir # Starting the Qt Application Vampir GUI |
||
</pre> |
</pre> |
||
| ⚫ | |||
'''Please note:''' |
|||
| ⚫ | |||
| ⚫ | |||
| ⚫ | |||
* As always performance is best, if Your trace files are available on a parallel filesystem, not on NFS. |
|||
== Running Vampir GUI with parallel VampirServer == |
== Running Vampir GUI with parallel VampirServer == |
||
Revision as of 15:37, 5 November 2020
| Description | Content |
|---|---|
| module load | devel/vampir |
| Availability | bwUniCluster | BwForCluster_Chemistry |
| License | [Vampir Professional License] |
| Citing | n/a |
| Links | Homepage | Tutorial | Use case |
| Graphical Interface | Yes |
| Included modules |
Introduction
Vampir and VampirServer are performance analysis tools developed at the Technical University of Dresden. With support from the Ministerium für Wissenschaft, Forschung und Kunst (MWK),
all Universities participating in bwHPC (see bwUniCluster_2.0) have acquired a five year license.
Versions and Availability
A list of versions currently available on all bwHPC Clusters can be obtained from the
Cluster Information System CIS
{{#widget:Iframe
| url=https://cis-hpc.uni-konstanz.de/prod.cis/bwUniCluster/devel/vampir
| width=99%
| height=120
}}
On the command line please check for availability using module avail devel/vampir. Vampir provides the GUI and allows analyzing traces of a few hundred Megabytes. For larger traces, you may want to revert to using a remote VampirServer running in parallel on the compute nodes via a Batch script (see below).
Application traces consist of information gathered on the clusters prior to running Vampir using VampirTrace or Score-P and include timing, MPI communication, MPI I/O, hardware performance counters and CUDA / OpenCL profiling (if enabled in the tracing library).
$ : bwUniCluster 2.0 $ module avail devel/vampir ------------------------ /opt/bwhpc/common/modulefiles/Core ------------------------- devel/vampir/9.9
Attention!
Do not run vampir on the head nodes with large traces with many analysis processes for a long period of time.
Please use on of the possibilities listed below.
Tutorial
For online documentation see the links section in the summary table at the top of this page. The local installation provides Manuals in the $VAMPIR_DOC_DIR directory.
Running Vampir GUI
Running Vampir GUI and VampirServer is possible in various way as highlighted in the following images:
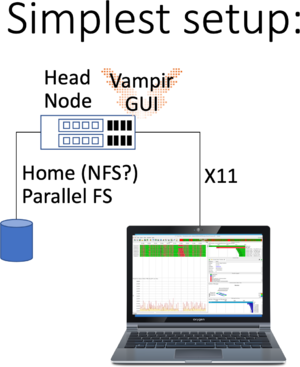
This is the simplest of all setups: You have to log in using X11 forwarding, by ticking the box in Putty, or passing -X or -Y to Your SSH line when logging in. Please check whether Your X11 forwarding has been setup and working by checking the $DISPLAY variable. For example run:
workstation$ ssh -Y user@bwunicluster.scc.kit.edu bwunicluster$ echo $DISPLAY 10.0.3.229:20.0 bwunicluster$ xdvi # If the window pops up, You're good to go bwunicluster$ module load devel/vampir bwunicluster$ vampir # Starting the Qt Application Vampir GUI
Please note:
- That You shouldn't run time-consuming, long-running tasks requiring lots of memory on the login nodes.
- Please see the net step of running additionally VampirServer
- As always performance is best, if Your trace files are available on a parallel filesystem, not on NFS.
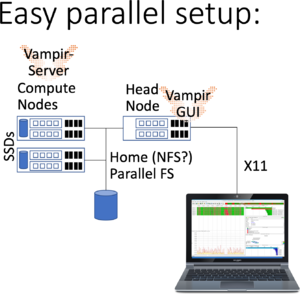
Running Vampir GUI with parallel VampirServer
In addition to the previous setup, You start a batch job to do the heavy lifting of analyzing large trace files employing the VampirServer.
bwunicluster$ module load devel/vampir bwunicluster$ sbatch --time=02:00:00 $VAMPIR_HOME/vampirserver.slurm Submitted batch job 1234
Please note that this submits a minimal batch allocation to queue multiple. Since this may take considerable amount to start (like 10 minutes to an hour), start another batch job but to the queue mainly aimed at testing and developping:
bwunicluster$ sbatch --partition=dev_multiple --time=30 $VAMPIR_HOME/vampirserver.slurm Submitted batch job 1235
The maximum amount of time is 30 minutes in this queue (on bwUniCluster check BwUniCluster_2.0_Batch_Queues). Please check squeue and the output file slurm-1234.out and slurm-1235.out for the nodes and the ports to connect to remotely. Then start vampir and connect to the nodes and the port -- and browse to your trace file. More information on VampirServer is provided in the below section.
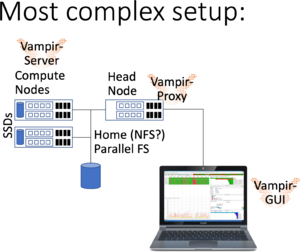
Running Vampir GUI locally with parallel VampirServer
The previous setups may have latency issues when connecting over slow network connections and VPN. An alternative is to use a vampir-proxy service on the head node and installing Vampir GUI locally on your work PC (available to bwHPC users employed at the partner Universities -- please contact your local compute center).
bwunicluster$ module load devel/vampir bwunicluster$ sbatch --time=02:00:00 $VAMPIR_HOME/vampirserver.slurm Submitted batch job 1234
Now, You need to start the vampir-proxy.
bwunicluster$ XXX
As always, there may be networking issues (VPN, Firewall, connectivity), which require assistance from your compute center.
Running remote VampirServer
The installation provides in $VAMPIR_HOME a SLURM batch script with which You may run parallel VampirServer instance on the compute nodes. You may attach to your VampirServer node using the provided port (typically port 30000, please check in the SLURM output file, once started). The SLURM script only supplies the queue name (default multiple); if You expect Your analysis to run for only 30 minutes or less, you may want to use another Queue meant for short-running development purposes dev_multiple which on bwUniCluster allows specifying the maximum time:
sbatch --partition=dev_multiple --time=30 $VAMPIR_HOME/vampirserver.slurm
Meanwhile, you may want to start another job using the default multiple queue; so that it will be scheduled, once your first job runs out.
Please query using squeue on the current status of both jobs and check the relevant SLURM output files.
VampirServer commands
If You want to run VampirServer as part of Your job-script, e.g. after finalizing Your application's run, add the following to your Batch script:
module load devel/vampir vampirserver start mpi
This shell scripts starts the MPI-parallel version of the VampirServer in the existing, already running SLURM job. The results of starting VampirServer is stored in $HOME/.vampir/server/list; you may check using the below commands, or by checking this file directly.
15 1604424211 mpi 20 uc2n001.localdomain 30000 2178460
Where the first column is the server number (incremented), the third column is the parallelisation mode VAMPIR_MODE, the next column is the number of tasks, followed by the name of the node (uc2n001) and the port (30000).
The commands available to the vampirserver shell script are
| Command | Description |
|---|---|
| help | Show this help |
| config | Interactively configure VampirServer for the given host system. |
| list | List running servers including hostname and port (see file $HOME/.vampir/server/list). |
| start [-t NUM] [LAUNCHER] | Starts a new VampirServer, using -t number of seconds with LAUNCHER being either smp (default), mpi and ap (Cray/HPE only) |
| stop [SERVER_ID] | Stops the specified server again |
| version | Print VampirServer's revision |