BwUniCluster2.0/Jupyter: Difference between revisions
| (60 intermediate revisions by 6 users not shown) | |||
| Line 17: | Line 17: | ||
= Access requirements = |
= Access requirements = |
||
{|style="background:#deffee; width:100%;" |
|||
To use Jupyter on the HPC resources of SCC, the access requirements for [https://wiki.bwhpc.de/e/BwUniCluster_2.0_User_Access bwUniCluster 2.0] apply. Registration at [https://bwidm.scc.kit.edu/ https://bwidm.scc.kit.edu/] is required. |
|||
|style="padding:5px; background:#cef2e0; text-align:left"| |
|||
[[Image:Attention.svg|center|25px]] |
|||
|style="padding:5px; background:#cef2e0; text-align:left"| |
|||
Access to Jupyter is '''limited to IP addresses from the BelWü network'''. |
|||
All home institutions of our current users are connected to BelWü, so if you are on your campus network (e.g. in your office or on the Campus WiFi) you should be able to connect to bwUniCluster 2.0 without restrictions. |
|||
If you are outside one of the BelWü networks (e.g. at home), a VPN connection to the home institution or a connection to an SSH jump host at the home institution must be established first. |
|||
|} |
|||
To use Jupyter on the HPC resources of SCC, the access requirements for [https://wiki.bwhpc.de/e/Registration/bwUniCluster bwUniCluster 2.0] apply. A [https://wiki.bwhpc.de/e/Registration/bwUniCluster registration] is required. Please note, You should've completed registration and tested your login once using [https://wiki.bwhpc.de/e/Registration/SSH Secure Shell (ssh)]. |
|||
The Jupyter service is only accessible from within the network of your home organization. To access it from outside, you must first establish a [https://wiki.bwhpc.de/e/BwUniCluster_2.0_User_Access_New#Establishing_network_access VPN Connection] to your home organization. |
|||
Currently, it is necessary to log in to the bwUniCluster once via SSH in order to use the Jupyter service. In the future, this step will be omitted. |
|||
= Login process = |
= Login process = |
||
Login takes place at |
|||
Login takes place at [https://uc2-jupyter.scc.kit.edu uc2-jupyter.scc.kit.edu]. For login, KIT username, KIT password and a 2-factor authentication is required. |
|||
* bwUniCluster 2.0: [https://uc2-jupyter.scc.kit.edu uc2-jupyter.scc.kit.edu] |
|||
* SDIL: [https://sdil-jupyter.scc.kit.edu sdil-jupyter.scc.kit.edu] |
|||
* HoreKa: [https://hk-jupyter.scc.kit.edu hk-jupyter.scc.kit.edu] |
|||
* HAICORE: [https://haicore-jupyter.scc.kit.edu haicore-jupyter.scc.kit.edu] |
|||
For login, your username, your password and a 2-factor authentication are required. |
|||
If you are not yet logged in to KIT, you will first be automatically redirected to the corresponding login page. Select your home organization (e.g. KIT) and press Continue. In the Login section that appears, enter your KIT username and password (not the service password). After pressing the Login button you will be redirected to the second factor query page. Enter the one-time password (e.g. from KIT Token or Google Authenticator App) and press Validate. |
|||
Now you will be redirected to the JupyterHub page, after pressing the "Sign in with your KIT Account" button you are logged in. |
|||
You will first find yourself on a landing page that also gives more information about the currently installed software versions. |
|||
[[File:anmeldung_750.gif|750px]] |
|||
By pressing the login button you will be redirected to the JupyterHub page. Click on Enter JupyterHub to start the login process. Select the organization (e.g. KIT) that has granted you access to the HPC system and press Continue. In the Login section that appears, enter your username and password (not the service password). |
|||
After pressing the Login button you will be redirected to the second factor query page. Enter the one-time password (e.g. from KIT Token or Google Authenticator App) and press Validate. Now you are done with the login process and can start selecting your computing resources. |
|||
[[File:Jupyter_Anmeldung.gif|700px]] |
|||
= Selection of the compute resources = |
= Selection of the compute resources = |
||
| Line 36: | Line 48: | ||
The Jupyter notebooks are executed in an interactive session on the compute nodes of the HPC clusters. Just like accessing an interactive session with SSH, resource allocation is done by the Workload Manager Slurm. The selection of resources for Jupyter is realized via drop-down menus. Only jobs with a maximum of one node are possible. |
The Jupyter notebooks are executed in an interactive session on the compute nodes of the HPC clusters. Just like accessing an interactive session with SSH, resource allocation is done by the Workload Manager Slurm. The selection of resources for Jupyter is realized via drop-down menus. Only jobs with a maximum of one node are possible. |
||
Available for selection are |
Available resources for selection are |
||
* Partition/Queue |
|||
* Runtime |
|||
* Number of GPUs |
|||
* Number of CPU cores |
* Number of CPU cores |
||
* Number of GPUs |
|||
* Runtime |
|||
* Partition/Queue |
|||
* Amount of main memory |
* Amount of main memory |
||
If Auto-Reservation is selected the automatic Jupyter reservation of the cluster is enabled. |
|||
In normal mode, the grayed-out fields contain reasonable presets, depending on the number of required CPU cores or GPUs respectively. The presets can be bypassed in advanced mode, where further options are available. |
|||
Advanced Mode can be activated by clicking on the checkbox of the same name. The following additional options then become available: |
|||
* Specification of a reservation |
|||
* LSDF mount option |
|||
* BEEOND mount option |
|||
After the selection is made, the interactive job is started with the spawn button. As when requesting interactive compute resources with the `salloc` command, waiting times may occur. These are usually the longer the larger the requested resources are. |
After the selection is made, the interactive job is started with the spawn button. As when requesting interactive compute resources with the `salloc` command, waiting times may occur. These are usually the longer the larger the requested resources are. |
||
Even if the chosen resources are available immediately, the spawning process may take up to one minute. |
|||
[[File:Ressources_neu.gif|500px]] |
|||
Please note that in advanced mode, resource combinations can be selected that are impossible to be met. In this case, an error message will appear when the job is spawned. |
|||
[[File:Jupyter_Falsche_ressourcen.gif|500px]] |
|||
The spawning timeout is currently set to 10 minutes. With a normal workload of the HPC facility, this time is usually sufficient to get interactive resources. |
|||
== Prioritized access to computing resources on bwUniCluster 2.0 == |
|||
The use of Jupyter requires the immediate availability of computing resources since the JupyterLab server is started within an interactive Slurm session. To improve the availability of CPUs/GPUs for interactive supercomputing with Jupyter, '''automatic reservation''' for CPU (single) and GPU (gpu_8) resources has been set up on '''bwUniCluster 2.0'''. It is active '''between 8am and 8pm''' every weekday. The reservation is automatically active if |
|||
* no other reservation is set manually |
|||
[[File:Ressourcen_750.gif|500px]] |
|||
* Auto-Reservation is enabled |
|||
To give you a better overview of the currently available resources, a status indicator has been implemented. It appears when selecting the number of required CPUs/GPUs and shows whether a Jupyter job of the selected size can currently be started or not. Green means the selected CPU/GPU resources are available instantly. Yellow means only a single additional job of the selected size can be started. Red means there are no GPU resources left that could satisfy the selected amount of resources. |
|||
If by mistake an impossible resource combination is selected, an error message is displayed. |
|||
If there are no more resources available within the reservation, you can try selecting a different amount of CPUs/GPUs or activate Advanced Mode and select a different partition. Availability can be estimated using sinfo_t_idle, which is available when logging in via SSH. |
|||
[[File:wrong_ressources_small.gif|500px]] |
|||
= JupyterLab = |
= JupyterLab = |
||
| Line 69: | Line 104: | ||
* Settings: general settings and an editor for advanced settings |
* Settings: general settings and an editor for advanced settings |
||
* Help: a list of help links to JupyterLab and the kernel |
* Help: a list of help links to JupyterLab and the kernel |
||
[[File:menu_small.gif|500px]] |
|||
== Left sidebar == |
== Left sidebar == |
||
| Line 80: | Line 114: | ||
* Tab Overview |
* Tab Overview |
||
* Lmod software selection: Search and load/unload Lmod software modules |
* Lmod software selection: Search and load/unload Lmod software modules |
||
[[File:seitenleiste_small.gif|750px]] |
|||
== Main working area == |
== Main working area == |
||
| Line 87: | Line 119: | ||
In a new JupyterLab session the Launcher tab is opened first. It contains buttons for starting new notebooks, code consoles and other functions. When a notebook is open, a new Launcher tab can be started by pressing the plus symbol in the file browser tab of the left sidebar, by calling ''File > New Launcher'' in the upper menu bar or by the key combination ''Ctrl+Shift+L''. |
In a new JupyterLab session the Launcher tab is opened first. It contains buttons for starting new notebooks, code consoles and other functions. When a notebook is open, a new Launcher tab can be started by pressing the plus symbol in the file browser tab of the left sidebar, by calling ''File > New Launcher'' in the upper menu bar or by the key combination ''Ctrl+Shift+L''. |
||
[[File:hauptbereich_small.gif|750px]] |
|||
== Classic Notebook == |
== Classic Notebook == |
||
| Line 134: | Line 164: | ||
== Software Stacks for Jupyter == |
== Software Stacks for Jupyter == |
||
Currently |
Currently 3 special Jupyter software stacks are available via Lmod: |
||
* jupyter/minimal |
|||
*: Minimal installation of JupyterLab |
|||
* jupyter/base |
* jupyter/base |
||
*: Basic installation of JupyterLab |
*: Basic installation of JupyterLab. |
||
*: For a complete list of pre-installed packages, please refer to [https://uc2-jupyter.scc.kit.edu/software-modules/#pre-installed-software-packages this site]. |
|||
* jupyter/tensorflow (default at login) |
* jupyter/tensorflow (default at login) |
||
*: Preinstalled software packages for machine learning applications. Includes among others TensorFlow, Keras, Torch, MXNet |
*: Preinstalled software packages for machine learning applications. Includes among others TensorFlow, Keras, Torch, MXNet, Pandas, Matplotlib, SKLearn. |
||
*: For a complete list of pre-installed packages and their respective version, please refer to [https://uc2-jupyter.scc.kit.edu/software-modules/#pre-installed-software-packages this site]. |
|||
* jupyter/extensions |
|||
Existing Python variants will be successively adapted for use in Jupyter. The integration of further programming languages and kernels is work in progress: '''Julia, R, C/C++''' (cling) |
|||
*: Same packages as tensorflow + extensions |
|||
These software stacks can be used both when accessing the cluster via JupyterHub, as well as for conventional access via SSH via module load. |
|||
A continuously updated list with the installed packages can be found on the corresponding subpage of the respective cluster: |
|||
* bwUniCluster 2.0: [https://uc2-jupyter.scc.kit.edu/software-modules uc2-jupyter.scc.kit.edu/software-modules] |
|||
* HoreKa: [https://hk-jupyter.scc.kit.edu/software-modules hk-jupyter.scc.kit.edu/software-modules] |
|||
= Installation of further software = |
= Installation of further software = |
||
The software provided by the Lmod modules jupyter/minimal, jupyter/base and jupyter/tensorflow can be easily supplemented by additional Python packages. There are 2 procedures for this. |
|||
<ul> |
|||
<li>User-Installation (not recommended)<br> |
|||
<code>pip install --user <packageName> </code><br> |
|||
The additional packages are installed under $HOME/.local/lib/python3.9/site-packages/ which is part of PYTHONPATH. |
|||
</li> |
|||
<li>Virtual environments (recommended)<br> |
|||
The user can create and use virtual environments (cf. Virtual environments). Packages provided by the jupyter Lmod modules remain visible and usable. |
|||
</li> |
|||
</ul> |
|||
== Virtual environments == |
|||
Python virtual environments allow to use different versions of a package and to keep your local site-packages (accessible under <code>$PYTHONPATH</code>) free from conflicts. |
|||
=== Creation of virtual environment === |
|||
<pre> |
|||
python -m venv <myEnv> |
|||
source <myEnv>/bin/activate |
|||
pip install <packageName> |
|||
deactivate |
|||
</pre> |
|||
The additional packages are installed under <code><myEnv>/lib/python3.9/site-packages/</code>. |
|||
=== Usage of virtual environment === |
|||
In order to use the virtual environment, it has to be activated via <code>source <myEnv>/bin/activate</code>. <code>PYTHONPATH</code> is set accordingly. Deactivation of the venv is done via <code>deactivate</code>. |
|||
=== Usage of virtual environment in JupyterLab === |
|||
To be able to use the virtual environments within JupyterLab, a corresponding kernel has to be installed: |
|||
<pre> |
|||
source <myEnv>/bin/activate |
|||
python -m ipykernel install \ |
|||
--user \ |
|||
--name myEnv \ |
|||
--display-name "Python (myEnv)" |
|||
</pre> |
|||
After installing the kernel (and possibly refreshing the browser window), a button named "myEnv" is available in JupyterLab. The kernel can also be selected from the drop-down menu. |
|||
'''Attention''' |
|||
The (Lmod) base module you used in the Creation of virtual environment step must be loaded to use the venv. However, to be on the safe side, you can also use the system Python (<code>/usr/bin/python3.9</code>) at creation time, which is available even without any <code>jupyter/{base,tensorflow}</code> module loaded. |
|||
== Examples on Data processing, Machine Learning & Visualization == |
|||
In the [https://github.com/hpcraink/workshop-parallel-jupyter/ workshop repository] the usage and best practices on Python in general, and the packages NumPy, Pandas, SciKit and Dask are provided, containing running examples based on open data. It also explains, how Jupyter interacts with pre-installed and your own provided environments. |
|||
== R language == |
|||
In order to use R language in JupyterLab, the Lmod module <code>math/R</code> has to be loaded (blue button in JupyterLab or <code>module add math/R</code> in terminal) and a corresponding kernel has to be installed. |
|||
<pre> |
|||
R |
|||
install.packages('IRkernel') |
|||
IRkernel::installspec() |
|||
</pre> |
|||
After installing the kernel , a button named "R" is available in JupyterLab. The kernel can also be selected from the drop-down menu. |
|||
'''Attention:''' |
|||
Don't forget to load the <code>math/R</code> module (blue button) before using the kernel. |
|||
== Julia language == |
|||
In order to use Julia language in JupyterLab, the Lmod module <code>devel/julia/1.6.2</code> has to be loaded (blue button in JupyterLab or <code>module devel/julia/1.6.2</code> in terminal) and a corresponding kernel has to be installed. |
|||
<pre> |
|||
julia |
|||
] |
|||
add IJulia |
|||
</pre> |
|||
After installing the kernel , a button named "Julia 1.6.2" is available in JupyterLab. The kernel can also be selected from the drop-down menu. |
|||
'''Attention:''' |
|||
Don't forget to load the <code>devel/julia/1.6.2</code> module (blue button) before using the kernel. |
|||
---- |
|||
= Use of own virtual environments = |
|||
Revision as of 17:14, 8 November 2024
Jupyter can be used as an alternative to accessing HPC resources via SSH. For this purpose only a web browser is required. Within the website source code of different programming languages can be edited and executed. Furthermore different user interfaces and terminals are available.
Short description of Jupyter
Jupyter is a web application, central component of Jupyter is the Jupyter Notebook. It is a document, which can contain formatted text, executable code sections and (interactive) visualizations (image, sound, video, 3D views).
The Jupyter notebooks are executed in an interactive session on the compute nodes of the respective cluster. Access is via any modern web browser. Data is prepared and visualized on the server and therefore does not have to be transmitted over the network. Only the resulting text, image, sound and video data is transmitted. Starting point of a Jupyter session is the HOME directory of the user on the respective cluster.
JupyterLab is a modern user interface, within which one or more Jupyter notebooks can be opened, edited and executed. The individual notebooks can be arranged as tabs or tiled. JupyterLab is the standard user interface. Besides JupyterLab the classic notebook user interface is available, in which only one Jupyter notebook per browser tab can be opened at a time.
A Jupyter Kernel describes a separate process, in which one Jupyter Notebook is executed at a time. Different kernels are available for different programming languages or language versions.
Before a Jupyter session is started, the access authorization must be checked first. This is done via JupyterHub, where the resources are selected, for example the number of CPU cores, GPUs or the required main memory.
A detailed documentation of the Jupyter project can be found at https://jupyter.readthedocs.io.
Access requirements
|
Access to Jupyter is limited to IP addresses from the BelWü network. All home institutions of our current users are connected to BelWü, so if you are on your campus network (e.g. in your office or on the Campus WiFi) you should be able to connect to bwUniCluster 2.0 without restrictions. If you are outside one of the BelWü networks (e.g. at home), a VPN connection to the home institution or a connection to an SSH jump host at the home institution must be established first. |
To use Jupyter on the HPC resources of SCC, the access requirements for bwUniCluster 2.0 apply. A registration is required. Please note, You should've completed registration and tested your login once using Secure Shell (ssh).
Login process
Login takes place at
- bwUniCluster 2.0: uc2-jupyter.scc.kit.edu
- SDIL: sdil-jupyter.scc.kit.edu
- HoreKa: hk-jupyter.scc.kit.edu
- HAICORE: haicore-jupyter.scc.kit.edu
For login, your username, your password and a 2-factor authentication are required.
You will first find yourself on a landing page that also gives more information about the currently installed software versions. By pressing the login button you will be redirected to the JupyterHub page. Click on Enter JupyterHub to start the login process. Select the organization (e.g. KIT) that has granted you access to the HPC system and press Continue. In the Login section that appears, enter your username and password (not the service password). After pressing the Login button you will be redirected to the second factor query page. Enter the one-time password (e.g. from KIT Token or Google Authenticator App) and press Validate. Now you are done with the login process and can start selecting your computing resources.
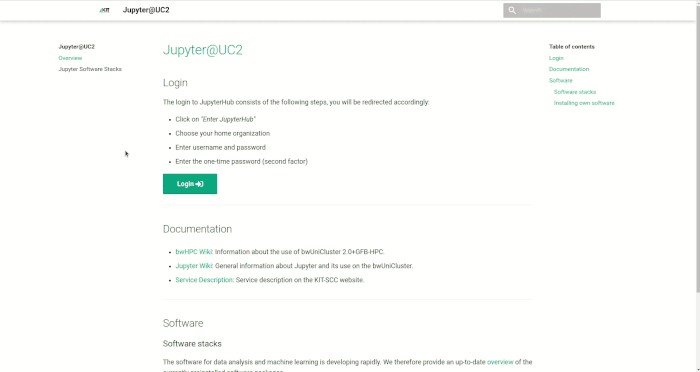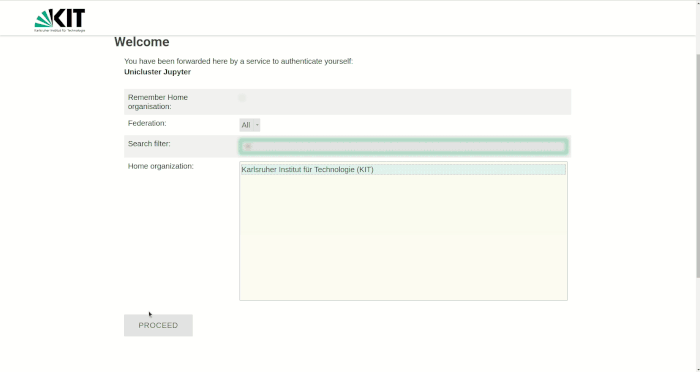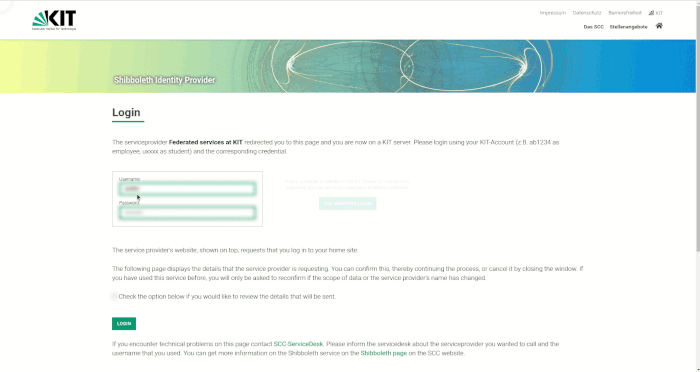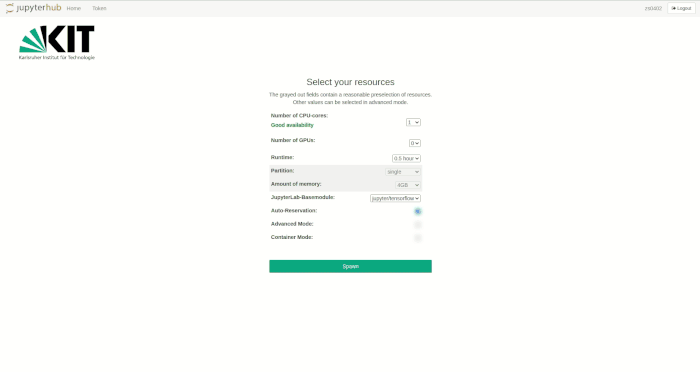
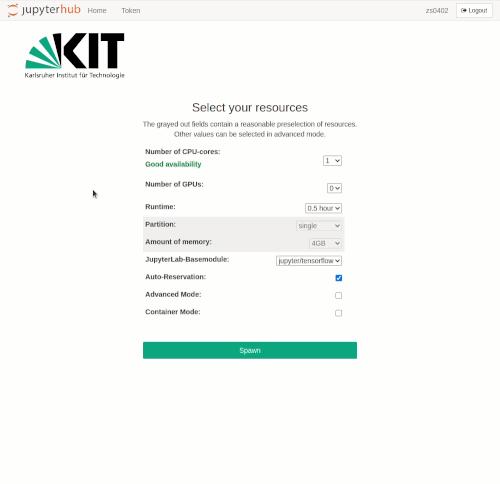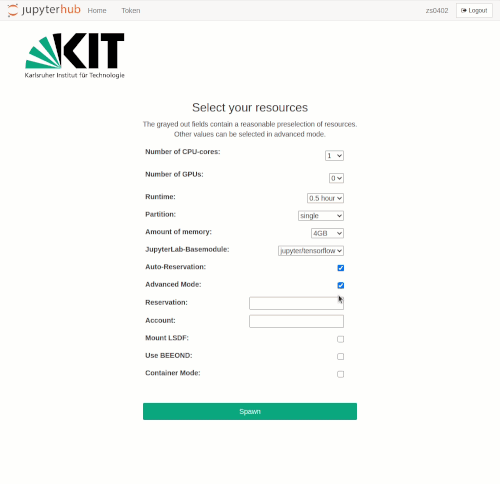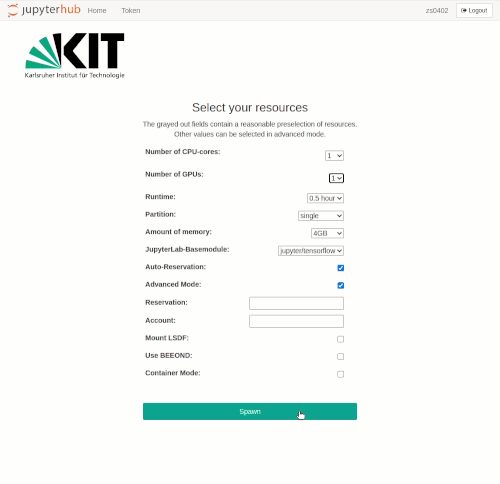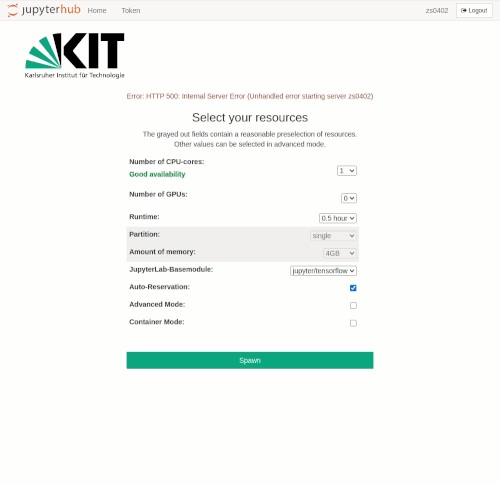
Selection of the compute resources
The Jupyter notebooks are executed in an interactive session on the compute nodes of the HPC clusters. Just like accessing an interactive session with SSH, resource allocation is done by the Workload Manager Slurm. The selection of resources for Jupyter is realized via drop-down menus. Only jobs with a maximum of one node are possible.
Available resources for selection are
- Number of CPU cores
- Number of GPUs
- Runtime
- Partition/Queue
- Amount of main memory
If Auto-Reservation is selected the automatic Jupyter reservation of the cluster is enabled.
In normal mode, the grayed-out fields contain reasonable presets, depending on the number of required CPU cores or GPUs respectively. The presets can be bypassed in advanced mode, where further options are available.
Advanced Mode can be activated by clicking on the checkbox of the same name. The following additional options then become available:
- Specification of a reservation
- LSDF mount option
- BEEOND mount option
After the selection is made, the interactive job is started with the spawn button. As when requesting interactive compute resources with the `salloc` command, waiting times may occur. These are usually the longer the larger the requested resources are. Even if the chosen resources are available immediately, the spawning process may take up to one minute.
Please note that in advanced mode, resource combinations can be selected that are impossible to be met. In this case, an error message will appear when the job is spawned.
The spawning timeout is currently set to 10 minutes. With a normal workload of the HPC facility, this time is usually sufficient to get interactive resources.
Prioritized access to computing resources on bwUniCluster 2.0
The use of Jupyter requires the immediate availability of computing resources since the JupyterLab server is started within an interactive Slurm session. To improve the availability of CPUs/GPUs for interactive supercomputing with Jupyter, automatic reservation for CPU (single) and GPU (gpu_8) resources has been set up on bwUniCluster 2.0. It is active between 8am and 8pm every weekday. The reservation is automatically active if
- no other reservation is set manually
- Auto-Reservation is enabled
To give you a better overview of the currently available resources, a status indicator has been implemented. It appears when selecting the number of required CPUs/GPUs and shows whether a Jupyter job of the selected size can currently be started or not. Green means the selected CPU/GPU resources are available instantly. Yellow means only a single additional job of the selected size can be started. Red means there are no GPU resources left that could satisfy the selected amount of resources.
If there are no more resources available within the reservation, you can try selecting a different amount of CPUs/GPUs or activate Advanced Mode and select a different partition. Availability can be estimated using sinfo_t_idle, which is available when logging in via SSH.
JupyterLab
JupyterLab is the standard user interface. In the following only its essential functions are briefly introduced. A detailed documentation is available at https://jupyterlab.readthedocs.io.
Menu bar
The menu bar at the upper edge of JupyterLab has higher-level menus that display the actions available in JupyterLab along with their shortcut keys. The default menus are:
- File: Actions related to files and directories
- Edit: Actions related to editing documents and other activities
- View: actions that change the appearance of JupyterLab
- Run: Actions to execute code in various activities like notebooks and code consoles
- Kernel: Actions to manage kernels that are separate processes for executing code
- Tabs: a list of open documents and activities in the Dock Panel
- Settings: general settings and an editor for advanced settings
- Help: a list of help links to JupyterLab and the kernel
Left sidebar
In the left sidebar there are foldable tabs. The most relevant ones are:
- File browser: Switch to directories and open files with left mouse button, context menu with right mouse button
- Running kernels: Overview of running kernels
- Command overview
- Tab Overview
- Lmod software selection: Search and load/unload Lmod software modules
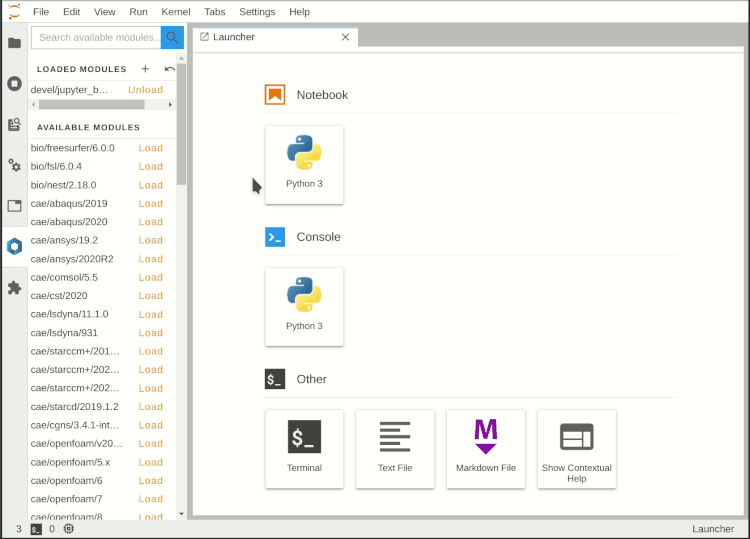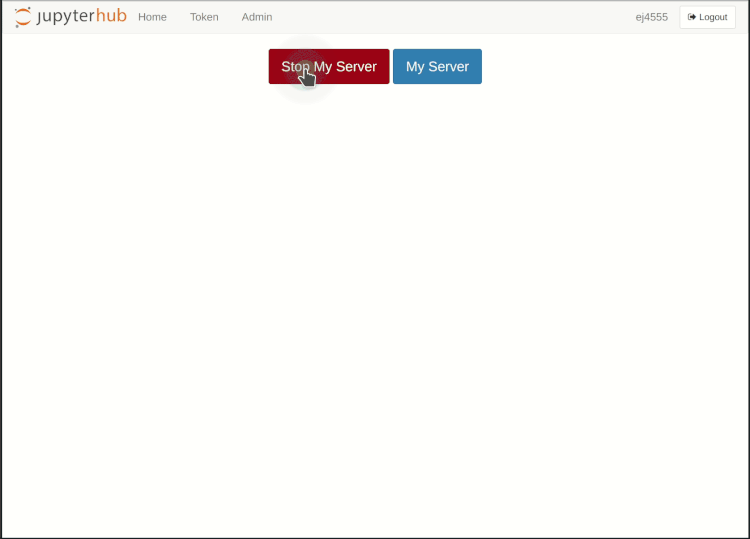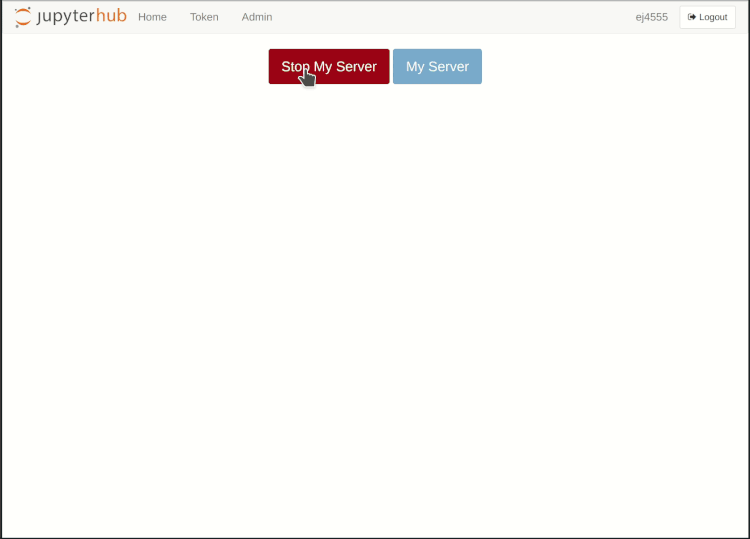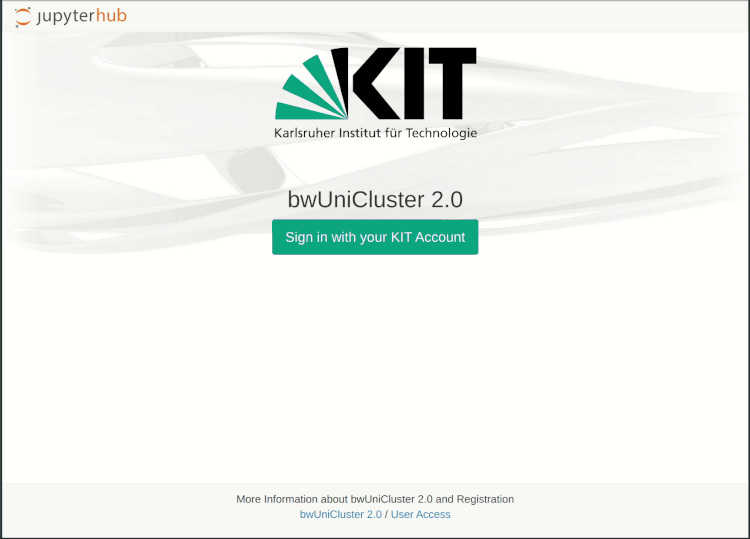
Main working area
The main working area in JupyterLab allows to arrange, resize and divide documents (notebooks, text files, etc.) and other activities (terminals, code consoles, etc.) in tabs. By holding down the left mouse button, the tabs can be grabbed and repositioned.
In a new JupyterLab session the Launcher tab is opened first. It contains buttons for starting new notebooks, code consoles and other functions. When a notebook is open, a new Launcher tab can be started by pressing the plus symbol in the file browser tab of the left sidebar, by calling File > New Launcher in the upper menu bar or by the key combination Ctrl+Shift+L.
Classic Notebook
The classic Jupyter Notebook user interface offers only one open Jupyter Notebook or terminal per browser tab. From the JupyterLab user interface the classic display can be reached in the menu bar under Help > Launch Classic Notebook. Clicking on the JupyterHub logo in the upper left corner will take you back to the JupyterLab interface.
Log out
You can log out from a running Jupyter session by calling File > Log Out in the upper menu bar.
|
As long as the interactive session is running, you can re-enter it at any time. Depending on the duration of your absence, it may be necessary to re-enter your one-time password and possibly KIT password.
If you want to end the interactive session before it has reached its runtime, you can do so via the Hub Control Panel. Under File > Hub Control Panel in the upper menu bar, it is opened in a new browser tab. By pressing the Stop My Server button the session will be terminated. You can now log out using the Logout button in the upper right corner or start a new session directly using the Start My Server button, for example with a changed resource selection.
Selection of software
For the selection of the required Lmod software modules the corresponding tab Softwares is available in the left sidebar. The list of available modules can be narrowed down by entering the search field. The desired module is loaded by pressing the Load button. In the list with the loaded modules you can remove them with the Unload button.
|
Software Stacks for Jupyter
Currently 3 special Jupyter software stacks are available via Lmod:
- jupyter/minimal
- Minimal installation of JupyterLab
- jupyter/base
- Basic installation of JupyterLab.
- For a complete list of pre-installed packages, please refer to this site.
- jupyter/tensorflow (default at login)
- Preinstalled software packages for machine learning applications. Includes among others TensorFlow, Keras, Torch, MXNet, Pandas, Matplotlib, SKLearn.
- For a complete list of pre-installed packages and their respective version, please refer to this site.
- jupyter/extensions
- Same packages as tensorflow + extensions
These software stacks can be used both when accessing the cluster via JupyterHub, as well as for conventional access via SSH via module load.
A continuously updated list with the installed packages can be found on the corresponding subpage of the respective cluster:
- bwUniCluster 2.0: uc2-jupyter.scc.kit.edu/software-modules
- HoreKa: hk-jupyter.scc.kit.edu/software-modules
Installation of further software
The software provided by the Lmod modules jupyter/minimal, jupyter/base and jupyter/tensorflow can be easily supplemented by additional Python packages. There are 2 procedures for this.
- User-Installation (not recommended)
pip install --user <packageName>
The additional packages are installed under $HOME/.local/lib/python3.9/site-packages/ which is part of PYTHONPATH. - Virtual environments (recommended)
The user can create and use virtual environments (cf. Virtual environments). Packages provided by the jupyter Lmod modules remain visible and usable.
Virtual environments
Python virtual environments allow to use different versions of a package and to keep your local site-packages (accessible under $PYTHONPATH) free from conflicts.
Creation of virtual environment
python -m venv <myEnv> source <myEnv>/bin/activate pip install <packageName> deactivate
The additional packages are installed under <myEnv>/lib/python3.9/site-packages/.
Usage of virtual environment
In order to use the virtual environment, it has to be activated via source <myEnv>/bin/activate. PYTHONPATH is set accordingly. Deactivation of the venv is done via deactivate.
Usage of virtual environment in JupyterLab
To be able to use the virtual environments within JupyterLab, a corresponding kernel has to be installed:
source <myEnv>/bin/activate
python -m ipykernel install \
--user \
--name myEnv \
--display-name "Python (myEnv)"
After installing the kernel (and possibly refreshing the browser window), a button named "myEnv" is available in JupyterLab. The kernel can also be selected from the drop-down menu.
Attention
The (Lmod) base module you used in the Creation of virtual environment step must be loaded to use the venv. However, to be on the safe side, you can also use the system Python (/usr/bin/python3.9) at creation time, which is available even without any jupyter/{base,tensorflow} module loaded.
Examples on Data processing, Machine Learning & Visualization
In the workshop repository the usage and best practices on Python in general, and the packages NumPy, Pandas, SciKit and Dask are provided, containing running examples based on open data. It also explains, how Jupyter interacts with pre-installed and your own provided environments.
R language
In order to use R language in JupyterLab, the Lmod module math/R has to be loaded (blue button in JupyterLab or module add math/R in terminal) and a corresponding kernel has to be installed.
R
install.packages('IRkernel')
IRkernel::installspec()
After installing the kernel , a button named "R" is available in JupyterLab. The kernel can also be selected from the drop-down menu.
Attention:
Don't forget to load the math/R module (blue button) before using the kernel.
Julia language
In order to use Julia language in JupyterLab, the Lmod module devel/julia/1.6.2 has to be loaded (blue button in JupyterLab or module devel/julia/1.6.2 in terminal) and a corresponding kernel has to be installed.
julia ] add IJulia
After installing the kernel , a button named "Julia 1.6.2" is available in JupyterLab. The kernel can also be selected from the drop-down menu.
Attention:
Don't forget to load the devel/julia/1.6.2 module (blue button) before using the kernel.