Helix/bwVisu/JupyterLab and SDS@hd/Access: Difference between pages
H Schumacher (talk | contribs) m (fixed kernel path) |
H Schumacher (talk | contribs) (Added troubleshooting section) |
||
| Line 1: | Line 1: | ||
This page provides an overview on how to access data served by SDS@hd. To get an introduction to data transfer in general, see [[Data_Transfer|data transfer]]. |
|||
[https://jupyter.org/ JupyterLab] is an integrated development environment (IDE) that provides a flexible and scalable interface for the Jupyter Notebook system. It supports interactive data science and scientific computing across over 40 programming languages (including Python, Julia, and R). |
|||
== |
== Prerequisites == |
||
* You need to be [[SDS@hd/Registration|registered]]. |
|||
The default python version can be seen by running <code>python --version</code> in the terminal. |
|||
* You need to be in the belwue-Network. This means you have to use the VPN Service of your HomeOrganization, if you want to access SDS@hd from outside the bwHPC-Clusters (e.g. via eduroam or from your personal notebook). |
|||
== Needed Information, independent of the chosen tool == |
|||
A different python version can be installed into a new virtual environment and then registered as IPython kernel for the usage in JupyterLab. This is explained in the chapter [[#Add_packages_via_conda_environments | add packages via conda environments]]. |
|||
* [[Registration/Login/Username| Username]]: Same as for the bwHPC Clusters |
|||
== Install Python Packages == |
|||
* Password: The Service Password that you set at bwServices in the [[SDS@hd/Registration|registration step]]. |
|||
* SV-Acronym: Use the lower case version of the acronym for all access options. |
|||
* Hostname: The hostname depends on the chosen network protocol: |
|||
** For [[Data_Transfer/SSHFS|SSHFS]] and [[Data_Transfer/SFTP|SFTP]]: ''lsdf02-sshfs.urz.uni-heidelberg.de'' |
|||
** For [[SDS@hd/Access/SMB|SMB]] and [[SDS@hd/Access/NFS|NFS]]: ''lsdf02.urz.uni-heidelberg.de'' |
|||
** For [[Data_Transfer/WebDAV|WebDAV]] the url is: ''https://lsdf02-webdav.urz.uni-heidelberg.de'' |
|||
== Recommended Setup == |
|||
Python packages can be added by installing them into a virtual environment and then creating an IPython kernel from the virtual environment. </br> |
|||
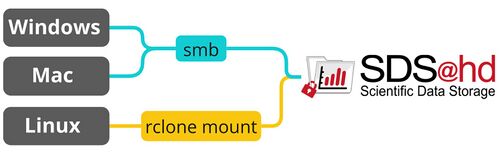
The following graphic shows the recommended way for accessing SDS@hd via Windows/Mac/Linux. The table provides an overview of the most important access options and links to the related pages.<br /> |
|||
<u>Kernels can be shared</u>. See the notes below. </br> |
|||
If you have various use cases, it is recommended to use [[Data_Transfer/Rclone|Rclone]]. You can copy, sync and mount with it. Thanks to its multithreading capability Rclone is a good fit for transferring big data.<br /> |
|||
If you want to move a virtual environment, it is adivsed to recreate it in the new place. Otherwise, dependencies based on relative paths will break. |
|||
For an overview of all connection possibilities, please have a look at [[Data_Transfer/All_Data_Transfer_Routes|all data transfer routes]]. |
|||
[[File:Data_transfer_diagram_simple.jpg|center|500px]] |
|||
# Create a virtual environment with... |
|||
<p style="text-align: center; font-size: small; margin-top: 10px">Figure 1: SDS@hd main transfer routes</p> |
|||
#* [[#Add_packages_via_venv_virtual_environments | ...venv]] or |
|||
#* [[#Add_packages_via_Conda_virtual_environments | ...conda]] (choose this option if you want to install a different python version) or |
|||
#* ...[https://docs.astral.sh/uv/getting-started/ uv] if you want to install a different python version but don't want to use conda. |
|||
# [[#Create_an_IPython_kernel | Create an IPython kernel]] from the virtual environment |
|||
# Use the kernel within JupyterLab |
|||
#* By default new kernels are saved under <code>~/.local/share/jupyter</code> and this location is automatically detected. Therefore, new kernels are directly available. |
|||
#* If the kernel is saved somewhere else, the path can be provided in the "Kernel path" field when submitting the job. For a kernel placed under <code>path_to_parent_dir/share/jupyter/kernels/my_kernel</code> the needed path would be <code>path_to_parent_dir/share/jupyter</code>. |
|||
{| class="wikitable" |
|||
<u>Notes regarding the sharing of IPython kernels</u> |
|||
|- style="font-weight:bold; text-align:center; vertical-align:middle;" |
|||
* The virtual environment and the kernel need to be placed in a shared directory. For example at SDS@hd. |
|||
! |
|||
* There could be a subdirectory for the virtual environments and one for the kernels. |
|||
! Use Case |
|||
* The path to the kernels is saved in the environment variable $JUPYTER_DATA_DIR. Jupyter relevant paths can be seen with <code>jupyter --paths</code> |
|||
! Windows |
|||
! Mac |
|||
! Linux |
|||
! Possible Bandwith |
|||
! Firewall Ports |
|||
|- |
|||
| [[Data_Transfer/Rclone|Rclone]] + <protocol> |
|||
| copy, sync and mount, multithreading |
|||
| ✓ |
|||
| ✓ |
|||
| ✓ |
|||
| depends on used protocol |
|||
| depends on used protocol |
|||
|- |
|||
| [[SDS@hd/Access/SMB|SMB]] |
|||
| mount as network drive in file explorer or usage via Rclone |
|||
| [[SDS@hd/Access/SMB#Windows|✓]] |
|||
| [[SDS@hd/Access/SMB#Mac|✓]] |
|||
| [[SDS@hd/Access/SMB#Linux|✓]] |
|||
| up to 40 Gbit/sec |
|||
| 139 (netbios), 135 (rpc), 445 (smb) |
|||
|- |
|||
| [[Data_Transfer/WebDAV|WebDAV]] |
|||
| go to solution for restricted networks |
|||
| [✓] |
|||
| ✓ |
|||
| ✓ |
|||
| up to 100GBit/sec |
|||
| 80 (http), 443 (https) |
|||
|- style="vertical-align:middle;" |
|||
| [[Data_Transfer/Graphical_Clients#MobaXterm|MobaXterm]] |
|||
| Graphical User Interface (GUI) |
|||
| [[Data_Transfer/Graphical_Clients#MobaXterm|✓]] |
|||
| ☓ |
|||
| ☓ |
|||
| see sftp |
|||
| see sftp |
|||
|- style="vertical-align:middle;" |
|||
| [[SDS@hd/Access/NFS|NFS]] |
|||
| mount for multi-user environments |
|||
| ☓ |
|||
| ☓ |
|||
| [[SDS@hd/Access/NFS|✓]] |
|||
| up to 40 Gbit/sec |
|||
| - |
|||
|- style="vertical-align:middle;" |
|||
| [[Data_Transfer/SSHFS|SSHFS]] |
|||
| mount, needs stable internet connection |
|||
| ☓ |
|||
| [[Data_Transfer/SSHFS#MacOS_&_Linux|✓]] |
|||
| [[Data_Transfer/SSHFS#MacOS_&_Linux|✓]] |
|||
| see sftp |
|||
| see sftp |
|||
|- style="vertical-align:middle;" |
|||
| [[Data_Transfer/SFTP|SFTP]] |
|||
| interactive shell, better usability when used together with Rclone |
|||
| [[Data_Transfer/SFTP#Windows|✓]] |
|||
| [[Data_Transfer/SFTP#MacOS_&_Linux|✓]] |
|||
| [[Data_Transfer/SFTP#MacOS_&_Linux|✓]] |
|||
| up to 40 Gbit/sec |
|||
| 22 (ssh) |
|||
|} |
|||
<p style="text-align: center; font-size: small; margin-top: 10px">Table 1: SDS@hd transfer routes</p> |
|||
=== |
=== Access from a bwHPC Cluster === |
||
More information about venv or other python virtual environments can be found at the [[Development/Python | Python]] page. |
|||
'''bwUniCluster'''<br /> |
|||
Steps for creating a '''venv''' virtual environment: |
|||
You can't mount to your $HOME directory but you can create a mount under $TMPDIR by following the instructions for [[Data_Transfer/Rclone#Usage_Rclone_Mount | Rclone mount]]. It is advised to wait a couple of seconds (<code>sleep 5</code>) before trying to use the mounted directory. |
|||
<ol style="list-style-type: decimal;"> |
|||
<li>Open a terminal.</li> |
|||
<li>Create a new virtual evironment: |
|||
<syntaxhighlight lang="bash">python3 -m venv <env_parent_dir>/<env_name></syntaxhighlight> |
|||
* <code>env_parent_dir</code> is the path to the folder where the virtual environment shall be created. Relative paths can be used. |
|||
* Caution: If you you want to share the environment with others, make sure to already create it in the shared place. |
|||
</li> |
|||
<li>Activate the environment: |
|||
<syntaxhighlight lang="bash">source <env_parent_dir>/<env_name>/bin/activate</syntaxhighlight></li> |
|||
<li>Update pip and install packages: |
|||
<syntaxhighlight lang="bash">pip install -U pip |
|||
pip install <packagename></syntaxhighlight></li> |
|||
<li> [[#Create_an_IPython_kernel | Create an IPython kernel]]</li> |
|||
</ol> |
|||
'''bwForCluster Helix'''<br /> |
|||
=== Add packages via Conda virtual environments === |
|||
You can directly access your storage space under ''/mnt/sds-hd/'' on all login and compute nodes. |
|||
More information about using conda can be found at the [[Development/Conda | Conda]] page. |
|||
'''bwForCluster BinAC'''<br /> |
|||
<ol style="list-style-type: decimal;"> |
|||
You can directly access your storage space under ''/mnt/sds-hd/'' on all login and compute nodes. The prerequisites are: |
|||
<li>Load the miniforge module by clicking first on the blue hexagon icon on the left-hand side of Jupyter's start page and then on the "load" button right of the entry for miniforge in the software module menu. |
|||
* The SV responsible has enabled the SV on BinAC once by writing to [mailto:sds-hd-support@urz.uni-heidelberg.de sds-hd-support@urz.uni-heidelberg.de] |
|||
<li>Open a terminal.</li> |
|||
* You have a valid kerberos ticket, which can be fetched with <code>kinit <userID></code> |
|||
<li>Create a new virtual environment: |
|||
* If you are the only person using the environment, you can install it in your home directory: |
|||
<syntaxhighlight lang="bash">conda create --name <env_name> python=<python version></syntaxhighlight> |
|||
* If you want to install it into a different directory, for example a shared place: |
|||
<syntaxhighlight lang="bash">conda create --prefix <path_to_shared_directory>/<env_name> python=<python version> |
|||
</syntaxhighlight> |
|||
</li> |
|||
<li>Activate your environment: |
|||
<syntaxhighlight lang="bash">conda activate <myenv></syntaxhighlight></li> |
|||
<li>Install your packages: |
|||
<syntaxhighlight lang="bash">conda install <mypackage></syntaxhighlight></li> |
|||
<li> [[#Create_an_IPython_kernel | Create an IPython kernel]]</li> |
|||
</ol> |
|||
'''Other'''<br /> |
|||
== Create an IPython Kernel == |
|||
You can mount your SDS@hd SV on the cluster yourself by using [[Data_Transfer/Rclone#Usage_Rclone_Mount | Rclone mount]]. As transfer protocol you can use WebDAV or sftp. For a full overview please have a look at [[Data_Transfer/All_Data_Transfer_Routes | All Data Transfer Routes]]. |
|||
=== Access via Webbrowser (read-only) === |
|||
Python kernels are implementations of the Jupyter notebook environment for different languages or virtual environments. You can switch between kernels easily, allowing you to use the best tool for a specific task. |
|||
conda_kernels |
|||
Visit [https://lsdf02-webdav.urz.uni-heidelberg.de/ lsdf02-webdav.urz.uni-heidelberg.de] and login with your SDS@hd username and service password. Here you can get an overview of the data in your "Speichervorhaben" and download single files. To be able to do more, like moving data, uploading new files, or downloading complete folders, a suitable client is needed as described above. |
|||
=== Create a kernel from a virtual environment === |
|||
== Best Practices == |
|||
<ol> |
|||
<li>Activate the virtual environment.</li> |
|||
<li>Install the ipykernel package: |
|||
<syntaxhighlight lang="bash">pip install ipykernel</syntaxhighlight></li> |
|||
<li>Register the virtual environment as custom kernel to Jupyter. |
|||
* If you are the only person using the environment: |
|||
<syntaxhighlight lang="bash">python3 -m ipykernel install --user --name=<kernel_name></syntaxhighlight> |
|||
The kernel can be found under <code>~/.local/share/jupyter/kernels/</code>. |
|||
* If you installed the environment in a shared place and want to have the kernel there as well: |
|||
<syntaxhighlight lang="bash">python3 -m ipykernel install --prefix <path_to_kernel_folder> --name=<kernel_name></syntaxhighlight> |
|||
The kernel can then be found under <code>path_to_kernel_folder/share/jupyter/kernels/<kernel_name></code>. As long as the same <code>path_to_kernel_folder</code> is used, all kernels will be saved next to each other in "kernels". |
|||
</li> |
|||
</ol> |
|||
* '''Managing access rights with ACLs''' <br /> -> Please set ACLs either via the [https://www.urz.uni-heidelberg.de/de/service-katalog/desktop-und-arbeitsplatz/windows-terminalserver Windows terminal server] or via bwForCluster Helix. ACL changes won't work when used locally on a mounted directory. |
|||
=== Multi-Language Support === |
|||
* '''Multiuser environment''' -<br /> -> Use [[SDS@hd/Access/NFS|NFS]] |
|||
== Troubleshooting == |
|||
JupyterLab supports over 40 programming languages including Python, R, Julia, and Scala. This is achieved through the use of different kernels. |
|||
* '''Issue:''' The credentials aren't accepted |
|||
==== R Kernel ==== |
|||
*: → '''[[Registration/Login/Username| Check your Username]]''' |
|||
*: → '''[[Registration/bwForCluster/Helix#Troubleshooting_with_the_Help_of_bwServices | Troubleshooting with bwServices]]''' |
|||
<ol style="list-style-type: decimal;"> |
|||
<li>On the cluster: |
|||
<pre>$ module load math/R |
|||
$ R |
|||
> install.packages('IRkernel')</pre></li> |
|||
<li>On bwVisu: |
|||
<ol style="list-style-type: decimal;"> |
|||
<li>Start Jupyter App</li> |
|||
<li>In left menu: load math/R</li> |
|||
<li>Open Console:</li> |
|||
<pre>$ R |
|||
> IRkernel::installspec(displayname = 'R 4.2')</pre> |
|||
<li>Start kernel 'R 4.2' as console or notebook</li> |
|||
</ol> |
|||
</ol> |
|||
==== Julia Kernel ==== |
|||
Load the math/julia module. Open the Terminal. |
|||
<pre> |
|||
julia |
|||
] |
|||
add IJulia |
|||
</pre> |
|||
After that, Julia is available as a kernel. |
|||
== Interactive Widgets == |
|||
JupyterLab supports interactive widgets that can create UI controls for interactive data visualization and manipulation within the notebooks. Example of using an interactive widget: |
|||
<pre class="{.python">from ipywidgets import IntSlider |
|||
slider = IntSlider() |
|||
display(slider)</pre> |
|||
These widgets can be sliders, dropdowns, buttons, etc., which can be connected to Python code running in the backend. |
|||
== FAQ == |
|||
<ol><li>'''My conda commands are interrupted with message 'Killed'.'''</br> |
|||
Request more memory when starting Jupyter.</li> |
|||
<li>'''How can I navigate to my SDS@hd folder in the file browser?'''</br> |
|||
Open a terminal and set a symbolic link to your SDS@hd folder in your home directory. For example:</li> |
|||
<pre>cd $HOME |
|||
mkdir sds-hd |
|||
cd sds-hd |
|||
ln -s /mnt/sds-hd/sd16a001 sd16a001</pre> |
|||
<li>'''Jupyterlab doesn't let me in but asks for a password.'''</br> |
|||
Try using more memory for the job. If this doesn't help, try using the inkognito mode of your browser as the browser cache might be the problem.</li> |
|||
<li>'''I prefer VSCode over JupyterLab. Can I start a JupyterLab job and then connect with it via VSCode?'''</br> |
|||
This is not possible. Please start the job directly on Helix instead. You can find the instructions at the [[Development/VS_Code#Connect_to_Remote_Jupyter_Kernel | VSCode page]].</li> |
|||
</ol> |
|||
Latest revision as of 11:07, 8 October 2025
This page provides an overview on how to access data served by SDS@hd. To get an introduction to data transfer in general, see data transfer.
Prerequisites
- You need to be registered.
- You need to be in the belwue-Network. This means you have to use the VPN Service of your HomeOrganization, if you want to access SDS@hd from outside the bwHPC-Clusters (e.g. via eduroam or from your personal notebook).
Needed Information, independent of the chosen tool
- Username: Same as for the bwHPC Clusters
- Password: The Service Password that you set at bwServices in the registration step.
- SV-Acronym: Use the lower case version of the acronym for all access options.
- Hostname: The hostname depends on the chosen network protocol:
Recommended Setup
The following graphic shows the recommended way for accessing SDS@hd via Windows/Mac/Linux. The table provides an overview of the most important access options and links to the related pages.
If you have various use cases, it is recommended to use Rclone. You can copy, sync and mount with it. Thanks to its multithreading capability Rclone is a good fit for transferring big data.
For an overview of all connection possibilities, please have a look at all data transfer routes.
Figure 1: SDS@hd main transfer routes
| Use Case | Windows | Mac | Linux | Possible Bandwith | Firewall Ports | |
|---|---|---|---|---|---|---|
| Rclone + <protocol> | copy, sync and mount, multithreading | ✓ | ✓ | ✓ | depends on used protocol | depends on used protocol |
| SMB | mount as network drive in file explorer or usage via Rclone | ✓ | ✓ | ✓ | up to 40 Gbit/sec | 139 (netbios), 135 (rpc), 445 (smb) |
| WebDAV | go to solution for restricted networks | [✓] | ✓ | ✓ | up to 100GBit/sec | 80 (http), 443 (https) |
| MobaXterm | Graphical User Interface (GUI) | ✓ | ☓ | ☓ | see sftp | see sftp |
| NFS | mount for multi-user environments | ☓ | ☓ | ✓ | up to 40 Gbit/sec | - |
| SSHFS | mount, needs stable internet connection | ☓ | ✓ | ✓ | see sftp | see sftp |
| SFTP | interactive shell, better usability when used together with Rclone | ✓ | ✓ | ✓ | up to 40 Gbit/sec | 22 (ssh) |
Table 1: SDS@hd transfer routes
Access from a bwHPC Cluster
bwUniCluster
You can't mount to your $HOME directory but you can create a mount under $TMPDIR by following the instructions for Rclone mount. It is advised to wait a couple of seconds (sleep 5) before trying to use the mounted directory.
bwForCluster Helix
You can directly access your storage space under /mnt/sds-hd/ on all login and compute nodes.
bwForCluster BinAC
You can directly access your storage space under /mnt/sds-hd/ on all login and compute nodes. The prerequisites are:
- The SV responsible has enabled the SV on BinAC once by writing to sds-hd-support@urz.uni-heidelberg.de
- You have a valid kerberos ticket, which can be fetched with
kinit <userID>
Other
You can mount your SDS@hd SV on the cluster yourself by using Rclone mount. As transfer protocol you can use WebDAV or sftp. For a full overview please have a look at All Data Transfer Routes.
Access via Webbrowser (read-only)
Visit lsdf02-webdav.urz.uni-heidelberg.de and login with your SDS@hd username and service password. Here you can get an overview of the data in your "Speichervorhaben" and download single files. To be able to do more, like moving data, uploading new files, or downloading complete folders, a suitable client is needed as described above.
Best Practices
- Managing access rights with ACLs
-> Please set ACLs either via the Windows terminal server or via bwForCluster Helix. ACL changes won't work when used locally on a mounted directory. - Multiuser environment -
-> Use NFS
Troubleshooting
- Issue: The credentials aren't accepted